Protein Design Games (Winter 2024)
A collaboration between Liberum Biotech and Rosetta Commons
Introduction
Designing and testing proteins has historically been a slow and arduous process. While recent computational advancements have made the design efforts considerably faster, real-world validation remains a bottleneck in many labs.
Through a collaboration between Liberum Biotech and Rosetta Commons, we are pleased to introduce the inaugural Winter Protein Design Games (Winter 2024)! We hope to enable a hackathon-style protein design workflow where your genius and creativity can be put to the test in a real-world setting.
The goal of these games is to encourage collaboration and learning between scientists working at the edge of this exciting field and to shine the spotlight on your contributions which push forward the frontiers of science. We are thrilled to have you take part in this competition.
With the generous help of the Commons, there will be a $5,000 USD prize awarded to the winning team. As well, you will be invited to summer RosettaCon to present your methodology for the winning design.
Background
In 1994, a tobacco etch virus (TEV) protease was reported as a tool for sequence-specific protein cleavage, enabling purification of tag-free proteins. In this demonstration, the sequence for TEVís cognate cleavage site (ENLYFQ/X where X is preferably a serine or glycine) was placed between a protein of interest and an affinity tag. The fusion protein was expressed and bound to affinity beads via the tag, and the protein of interest was then cleaved off using TEV protease, releasing the tag-free target protein. The TEV protein itself, already fused with a different affinity tag (i.e. a 6x histidine tag), was then removed through incubation of the flow-through with Ni-NTA beads.
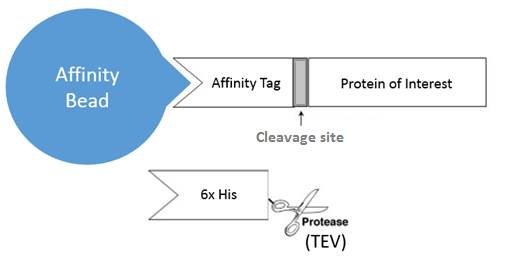
Due to its high specificity, efficiency at low temperatures and tolerance to a wide range of buffer conditions TEV protease has become the workhorse of protein biology labs. Today, the most common variant of TEV used is the 27 kDa catalytic domain with the S219V mutation discovered by the Waugh lab.
The ability to express functional TEV tagged with a 6xHis tag at its N-terminus has already been demonstrated in our bacterial cell-free system. Our cell-free system contains all the enzymes required for transcription and translation of the protein starting from DNA containing the constructís coding sequence.
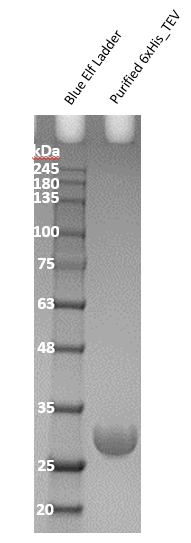
>6xHis_TEV
MHHHHHHGESLFKGPRDYNPISSTICHLTNESDGHTTSLYGIGFGPFIITNKHLFRRNNGTLLVQSLHGVFKVKNTTTLQQHLIDGRDMIIIRMPKDFPPFPQKLKFREPQREERICLVTTNFQTKSMSSMVSDTSCTFPSSDGIFWKHWIQTKDGQCGSPLVSTRDGFIVGIHSASNFTNTNNYFTSVPKNFMELLTNQEAQQWVSGWRLNADSVLWGGHKVFMSKPEEPFQPVKEATQLMN
>TEV_no_tag
MGESLFKGPRDYNPISSTICHLTNESDGHTTSLYGIGFGPFIITNKHLFRRNNGTLLVQSLHGVFKVKNTTTLQQHLIDGRDMIIIRMPKDFPPFPQKLKFREPQREERICLVTTNFQTKSMSSMVSDTSCTFPSSDGIFWKHWIQTKDGQCGSPLVSTRDGFIVGIHSASNFTNTNNYFTSVPKNFMELLTNQEAQQWVSGWRLNADSVLWGGHKVFMSKPEEPFQPVKEATQLMN
∑ Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Release of Proteins and Peptides from Fusion Proteins Using a Recombinant Plant Virus Proteinase. Anal Biochem. 1994;216(2):413Ė7. 10.1006/abio.1994.1060
∑ DarÚs JA, Schaad MC, Carrington JC. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J Virol. 1999;73(10):8732Ė40.
∑ Kapust RB, TŲzsťr J, Copeland TD, Waugh DS. The P1′ specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294(5):949Ė55. 10.1016/S0006-291X(02)00574-0
∑ Wu X, Wu D, Lu Z, Chen W, Hu X, Ding Y. A novel method for high-level production of TEV protease by superfolder GFP tag. J Biomed Biotechnol. 2009;2009:591923. 10.1155/2009/591923
Objective
Your teamís objective is to come up with a TEV protease design that outperforms wild-type in cleavage activity of a substrate. Your constructs will be synthesized, amplified with PCR and dropped directly into our cell-free protein synthesis system without PCR product purification. The crude reaction will be directly assayed using a fluorophore/quencher style TEV cleavage assay at 22C for 30 minutes.
Hints and Thoughts
Some PDB versions of the TEV protease contain truncations that may affect activity. Please make sure to take this into consideration when finalizing your designs. For example 1LVM contains a truncation at the C-terminus; while we do not know the effect of this particular truncation on activity, given the fact that self-cleavage near the C-terminus is known to affect activity, you may want to give some thought to including the missing parts in your designs.
Most commercially available TEV proteases seem to contain the S219V mutation that reduces self-cleavage and thus improves overall activity. You are not necessarily obligated to use this mutation.
TEV protease is often expressed fused to an N-terminal maltose binding protein (MBP) domain which increases solubility and is subsequently cleaved off by the TEV protease. The addition of MBP to the N-terminus is not allowed for the purposes of this competition, nor is it necessary.
Since activity of your TEV protease will be judged from crude, unpurified cell-free reactions you must balance both catalytic activity AND solubility in an environment resembling E. coli cytosol.
If you choose to optimize the nucleotide sequence of your constructs then you must accept the risk and consequences of doing so. We recommend you leave the nucleotide optimization to us and simply submit the amino acid sequence.
To date there have been over a dozen mutations to TEV protease published in the literature. Your teamís strategy may include a careful curation of the available non-patented mutations. While using known mutations may be safer, novel mutations or de novo designs may yield greater rewards.
The Rules
1) Your designs may include any of the following:
i) de novo designs (Does not have to have resemblance to TEV protease)
ii) Novel mutations to the target (via new or existing models or rational design)
iii) A curation of the mutations already available in the literature
2) Designs should be limited to the coding sequence alone; you will not be permitted to provide modifications to regulatory sequences (e.g. the promoter).
3) Constructs must include a 6xHis tag somewhere in the sequence. This can be N-terminal, C-terminal or anywhere else.
4) Your constructs cannot be longer than 267 amino acids long, including the 6xHis tag and the start codon.
5) There is space for up to 8 teams to submit 3 designs per team. If less than 8 teams sign up then remaining design space will be distributed between the teams (i.e. 6 teams with 4 designs each).
6) Each team must include at least two people from at least two different institutions. You must form your team at the conference and add your names to the sign-up sheet.
7) Deadline for submission is March 22, 2024 at midnight PT .
Evaluation Criteria
Each team will be scored out of a total of 10 points:
7/10 Points: Activity of the teamís best design when expressed in E. coli cell-free judged by cleavage of the canonical substrate ENLYFQ/G in a fluorophore/quencher based assay
∑ If the activity is less than wild-type, zero points are given
∑ The fluorescence of your teamís best design will be divided by the fluorescence of the competitions best design (or our positive control, whichever is higher) and multiplied by 7
∑ e.g. If your design produced 50,000 RFUs while the best design produced 70,000 RFUs, you will be awarded 5/7 points
3/10 Points: Communicating your methodology
∑ 1 Point for Brevity: Is your write up brief and concise?
∑ 1 Point for Replicability: Does your write-up include enough information to be replicated by the community, including all references in literature and code used?
∑ 1 Point for Aesthetics: Does your write up look presentable to the community?
Submission Instructions
Sequences and write-ups must be submitted to aidan@liberumbio.com before the designated deadline.
Sequences must be placed in a single excel document. Please ensure all sequences begin with a start codon (i.e. ATG or M). Kindly place all sequences in column A with no additional rows before or in between.
All write-up must be submitted in html format (with all the relevant dependencies such as css files, etc). If the original write-up it in MS-Word, please save it as a Web Page and submit the html file and associated folder as a zip file. This way, we just post your write-ups on our server with your original designs/layouts.
Include all team membersí names and email addresses for contact purposes
Terms
1) As participants, you attest that the sequences were generated without knowingly infringing any third party's intellectual property rights
2) You confirm that the designs are bio-safe, and that no alterations have been made to the target to make them toxic or pathogenic in any way
3) You acknowledge that while experiments will be conducted in a professional manner and judged as fairly as possible, characterization and timelines will not be perfect. You also agree that all results are final and will not be subject to reinvestigation
4) You agree that any work submitted here will be in the public domain and accessible to everyone
